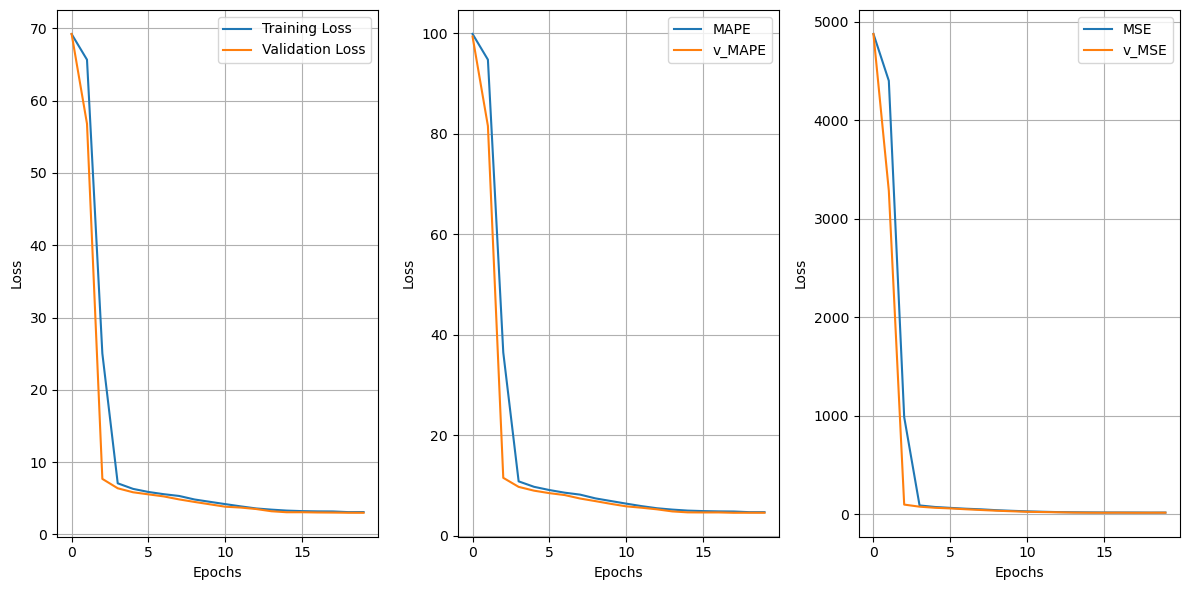
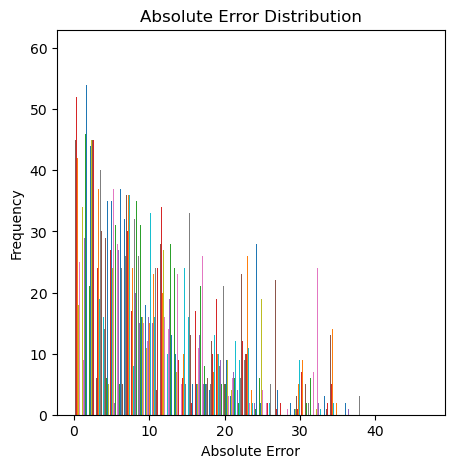
array([63.28493162, 64.27769707, 64.24859625, 75.27033822, 75.09546198,
75.09455566, 75.67042168, 75.5674344 , 64.72692714, 64.79060448,
73.62910063, 73.65050985, 79.21344943, 79.23629543, 72.82693221,
72.88084677, 86.10000331, 85.86183729, 79.56071386, 79.132046 ,
72.32717225, 69.03080151, 72.75008102, 72.78604679, 75.84212379,
75.8650568 , 67.45418302, 67.26566414, 75.61516906, 75.3735592 ,
76.54800706, 76.38556271, 80.84777011, 80.89375474, 69.69366366,
72.42435932, 66.31216642, 65.61576181, 70.08576218, 69.9833968 ,
74.48122982, 74.20373587, 74.16342442, 74.25302967, 70.37938279,
69.99105372, 76.58132703, 75.3147985 , 73.36448854, 75.28235669,
78.1310823 , 77.99838215, 62.64991602, 61.56493012, 70.872569 ,
72.43673557, 68.42888699, 66.82594233, 64.97802217, 64.80220869,
77.53687864, 77.48479044, 57.63558552, 55.83182247, 57.17589289,
57.08545012, 77.78829449, 74.67105451, 69.22266128, 73.57817612,
67.88597507, 67.55055629, 62.03831576, 66.4800068 , 74.44498136,
74.35477112, 78.84277519, 78.57979455, 71.6660567 , 75.81781643,
78.49059565, 77.43913431, 84.41378739, 83.37668892, 61.11522541,
60.92975373, 71.17473782, 72.67477566, 73.84684464, 73.21170279,
71.51480137, 74.44850268, 73.57589434, 73.44234334, 58.62753644,
56.38040847, 60.09866227, 60.11637266, 78.36257985, 78.39457351,
77.06726094, 76.76396422, 78.67210514, 78.6454721 , 77.43905014,
77.5072683 , 66.28674908, 67.47671129, 65.02979872, 65.0881979 ,
73.4490736 , 73.56486948, 81.2270811 , 81.07798155, 69.34626657,
65.8439691 , 79.39576605, 79.21577675, 75.74669664, 76.13209337,
68.47462227, 69.4284191 , 62.02441871, 62.47129517, 63.82243698,
62.91266769, 68.5682732 , 68.55293412, 64.97910191, 65.41127171,
68.18636685, 71.04329185, 81.07887315, 79.52767157, 75.9714915 ,
80.83334805, 74.49902042, 70.8401859 , 70.57013197, 72.76382284,
76.3707057 , 76.41857878, 68.31154669, 67.15413451, 83.68576647,
83.64783531, 77.92962151, 77.82397086, 80.87711054, 81.1979738 ,
72.57648043, 72.30601381, 78.4167785 , 77.7683826 , 74.36522602,
74.32762524, 75.74034336, 75.77161573, 66.47520823, 65.07594091,
72.66080086, 73.62678216, 70.71293595, 71.2146239 , 72.59992122,
72.22583114, 66.5251915 , 66.08054518, 73.32323065, 73.28699871,
60.97824448, 60.79887415, 62.89264576, 65.48090617, 74.41298198,
74.49792487, 80.22905801, 80.07087462, 74.34057063, 74.32616412,
65.21120056, 66.02124183, 64.07825827, 63.68915426, 72.54506289,
72.22861364, 70.61863412, 70.24914773, 61.47598447, 59.37804188,
78.6531124 , 78.48408632, 62.97346204, 59.47729302, 74.37728622,
74.3818426 , 73.76290316, 73.24985114, 71.2718747 , 71.50463038,
76.07162079, 73.19183159, 76.50410476, 73.47453547, 72.70717374,
72.6456079 , 62.69643707, 61.78537158, 65.53123655, 64.59679335,
68.31673613, 68.10024555, 69.68782951, 69.44226371, 84.09026014,
83.02687603, 84.58032396, 84.62719607, 71.39413139, 71.2497909 ,
57.07433471, 57.212554 , 55.03024141, 53.58639062, 79.3429749 ,
79.10816197, 74.39877515, 74.35617183, 65.9489262 , 65.75825716,
67.93438432, 69.54888002, 71.62850789, 69.57556582, 73.59670618,
72.86434158, 68.82939864, 69.75723616, 80.73075505, 76.16550381,
78.15151872, 77.43537566, 74.62927564, 74.86715198, 79.1350676 ,
78.07372115, 75.79868318, 75.30891527, 67.28081736, 67.08158799,
72.45679027, 72.38566758, 73.24053627, 73.28153695, 72.64336739,
72.8636013 , 67.61071872, 69.07396875, 77.88367647, 77.16781725,
65.38176969, 65.0393611 , 74.90250319, 74.78071558, 72.82889369,
73.11810039, 65.39023278, 65.41898819, 78.44226457, 78.50508405,
78.80680048, 78.77654688, 82.03745066, 81.01581952, 68.58547716,
68.74020574, 66.75074668, 68.24725704, 83.48502883, 82.42010017,
72.31260616, 71.97761959, 71.94697922, 71.97956388, 65.34583458,
63.67509382, 79.94831882, 79.92443263, 79.74146321, 79.57345789,
65.41387719, 69.33288008, 70.48249548, 70.34294422, 72.49512271,
72.5296839 , 73.63167428, 73.74922574, 69.83907307, 69.49706754,
68.83035928, 68.44598107, 73.33018822, 75.89818716, 71.56709598,
71.52544512, 72.7699083 , 75.89432363, 76.15421848, 76.12622032,
70.33586196, 70.17163129, 63.8257878 , 63.11621233, 69.18342035,
71.59013369, 74.39637723, 71.06658623, 76.83424233, 76.75897656,
72.20907206, 72.0332648 , 68.77446201, 68.77730286, 74.2745724 ,
74.45026269, 69.66084003, 68.07193834, 63.28810188, 65.3195207 ,
67.08485043, 66.21657961, 63.35835326, 61.79454959])